TCR Discovery Platform
Dr. Michael Volkmar
Aim
At the TCR Discovery Platform, our expertise is the identification, isolation and cloning of cancer-specific T cell receptor candidates. We are also developing methods for comprehensive functional characterization of TCRs.
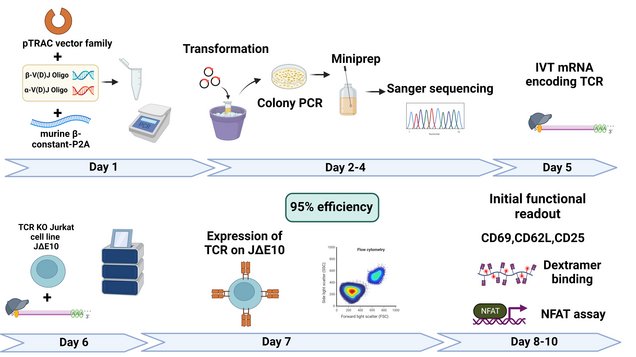
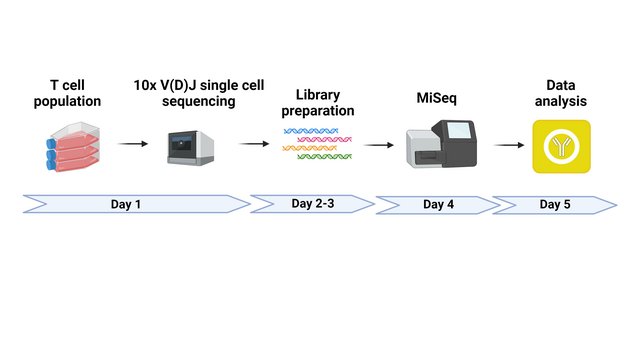
The main method we use to identify TCRs is single cell transcriptomics (scRNA-seq), but we also apply more traditional methods such as 5’RACE. Once TCR chain pairs have been identified, we clone them using a highly efficient cloning system (see publications) that we constantly improve and expand to accommodate collaborators’ needs. For example, we regularly achieve transfection rates of ≥95% with in vitro-transcribed RNA from TCR cDNAs (TCR IVT-RNA) but our TCR vector system also encompasses plasmid (pcDNA3.1 backbone), viral and transposon vectors. Cloning of TCRs into collaborators’ vectors is, naturally, also possible.
When tasked with functional characterization of candidate TCRs, we use our Jurkat E6.1-derived reporter cell line to assess proper cell surface expression of the TCR, avidity of TCR-transfected T cells to their target cells, and initiation of TCR signaling upon T cell-target cell contact.
While we also conduct own research, our primary focus is scientific collaboration projects to which we contribute our TCR- and T cell-related knowledge. Please feel free to contact us!
To stay competitive in the rapidly evolving field of TCR analysis we closely work with partners in the HI-TRON founding institutions in Heidelberg and Mainz.
Main techniques:
● Single cell RNA sequencing
(10X gene expression, VDJ, surface markers)
● TCR cloning
(plasmid, viral and transposon vectors, IVT-RNA)
● TCR-transgenic T cell avidity analysis
● NGS data analasis
(whole genome/exome seq, RNA-seq)
● HLA typing on basis of NGS data
● Neoepitope prediction and cloning
Methods under development:
● tumor dissociation and cryopreservation
●TIL (tumor-infiltrating lymphocyte) expansion culture
● MLTC (mixed lymphocyte-tumor culture)
● T cell enrichment (MACS, FACS)
● live cell imaging
● digital droplet PCR
(e.g. mutational burden in circulating tumor DNA
from blood plasma, CNV analyses of transgenes
from genomic DNA, …)
● Immunohistochemistry
Devices:
● 10X Genomics Chromium Controller
● Illumina MiSeq
● Agilent TapeStation 4200
● Bio-Rad QX200 Digital Droplet PCR,
CFX Opus 96 Real-time PCR System
● Lonza 4D-Nucleofector
● CTL S6 Ultra Immunospot Analyzer
● Sartorius Incucyte S3 Live Cell Analysis Instrument
TCR Discovery Platform
Dr. rer. nat. Michael Volkmar
 J. Arlt
J. Arlt
[since 2020] Head of TCR Discovery Platform, HI-TRON Mainz
[2012 - 2020] Senior Postdoctoral Fellow & Head of Project DKFZ. Heidelberg, Dept. Molecular Oncology of Gastrointestinal Tumors.
[2008 - 2012] Postdoc Université Libre de Bruxelles, Laboratory of Cancer Epigenetics.
[2002 - 2008] Doctorate, Institute for Genetics, Dept. Developmental Genetics.
HI-TRON Mainz DKFZ
TCR Discovery Platform
Obere Zahlbacherstr. 63
Building 911
55131 Mainz
Team
Selected publications
1. Volkmar M, Fakhr E, Zens S, Offringa R, Bury A, Gordon J, Huduti E, Wölfel T, Wölfel C. Identification of TRDV-TRAJ V-domains in human and mouse T cell receptor (TR) repertoires. accepted for publication in Frontiers in Immunology.
2. Meng Z, Rodriguez Ehrenfried A, Tan CL, Steffens LK, Kehm H, Zens S, Lauenstein C, Paul A, Schwab M, Förster JD, Salek M, Riemer AB, Wu H, Eckert C, Leonhardt C, Strobel O, Volkmar M, Poschke I, Offringa R. Transcriptome-based dissection of the tumor-reactive and bystander CD8+ T-cell repertoires in human pancreatic cancer. accepted for publication in Science Translational Medicine.
3. Nedwed AS, Helbich SS, Braband KL, Volkmar M, Delacher M, Marini F. Using combined single-cell gene expression, TCR sequencing and cell surface protein barcoding to characterize and track CD4 T cell clones from murine tissues. accepted for publication in Frontiers in Immunology.
4. Neulinger-Muñoz M, Schaack D, Grekova SP, Bauer AS, Giese T, Salg GA, Espinet E, Leuchs B, Heller A, Nüesch JPF, Schenk M, Volkmar M, Giese N. Human Retrotransposons and the Global Shutdown of Homeostatic Innate Immunity by Oncolytic Parvovirus H-1PV in Pancreatic Cancer. Viruses. 2021; 13(6):1019.
5. Kropp KN, Schäufele TJ, Fatho M, Volkmar M, Conradi R, Theobald M, Wölfel T, Catherine Wölfel C. A bicistronic vector backbone for rapid seamless cloning and chimerization of αβT-cell receptor sequences. PLoS ONE. 2020; 15(9):e0238875.
6. Zurli V, Montecchi T, Heilig R, Poschke I, Volkmar M, Wimmer G, Boncompagni G, Turacchio G, D’Elios MM, Campoccia G, Resta N, Offringa R, Fischer R, Acuto O, Kabanova A. Phosphoproteomics of CD2 signaling reveals AMPK-dependent regulation of lytic granule polarization in cytotoxic T cells. Sci Signal. 2020; 13(631):eaaz1965.
7. Wickström S, Lövgren T, Volkmar M, Reinhold BB, Duke-Cohan JS, Hartmann L, Rebmann J, Mueller A, Melief J, Maas RR, Ligtenberg M, Hansson J, Offringa R, Seliger B, Kiessling R. Cancer Neoepitopes for Immunotherapy: Discordance Between Tumor-Infiltrating T Cell Reactivity and Tumor MHC Peptidome Display. Front Immunol. 2019; 10:02766.
8. König AK, Fritz S, Volkmar M, Tampakis A, Youm J, Gaida MM, Werner J, Hackert T, Büchler MW, Offringa R, Strobel O. Mutational profile in IPMN subtypes. Pancreatology. 2017; 17(3): S25.
Platform News
10/2023: New members join the TCR Platform: Anna Gerstner and Maria Wiegand.